Global Genome NER
Global genome nucleotide excision repair (GG-NER) detects and eliminates bulky damages in the entire genome, including the untranscribed regions and silent chromatin (active and inactive genes throughout the genome), and this process does not depends on transcription. This process is controlled by XPC, a specialized protein factor that reveals the damage.
The pathway employs several "damage sensing" proteins including the DNA-damage binding (DDB) and XPC-Rad23B complexes that constantly scan the genome and recognize helix distortions; the XPC-Rad23B complex is responsible for distortion recognition, while DDB1 and DDB2 (XPE) can also recognize some types of damage caused by UV light. Upon identification of a damaged site, subsequent repair proteins are then recruited to the damaged DNA to verify presence of DNA damage, excise the damaged DNA surrounding the lesion then fill in the repair patch.
Damage Recognition
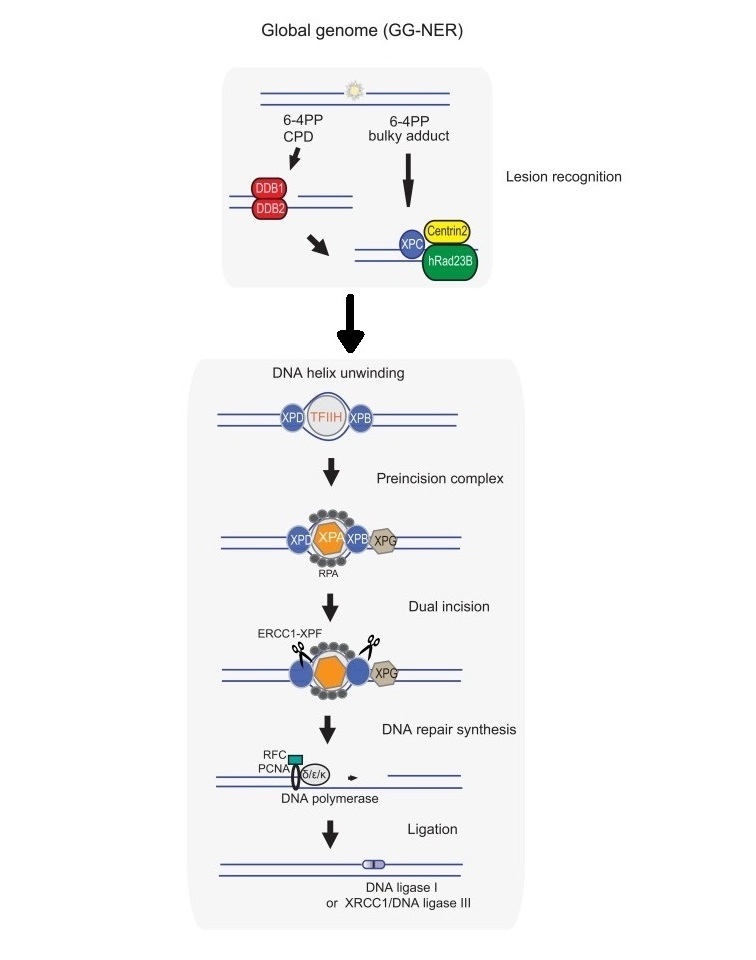
Schematic representation for mechanisms of global genome nucleotide excision repair (GG-NER) in mammals.
In GG-NER subpathway, the damage sensor XPC, in complex with UV excision repair protein RAD23 homologue B (RAD23B) and centrin 2 (CETN2), constantly probes the DNA for helix-distorting lesions, which are recognized with the help of the UV–DDB (ultraviolet (UV) radiation–DNA damage-binding protein) complex. Upon binding of the XPC complex to the damage, RAD23B dissociates from the complex. After damage recognition, the TFIIH (transcription initiation factor IIH) complex is recruited to the lesion in GG-NER. In NER the XPG structure - specific endonuclease, either associated with TFIIH or separately, binds to the pre-incision NER complex. Upon binding of TFIIH, the CAK (CDK-activating kinase) subcomplex dissociates from the core TFIIH complex. The helicase activity of TFIIH further opens the double helix around the lesion, and 5′–3′ unwinding of the DNA by the TFIIH basal transcription factor complex helicase subunit XPD verifies the existence of lesions with the help of the ATPase activity of the TFIIH XPB subunit and XPA, which binds to single-stranded, chemically altered nucleotides. In this step the single-stranded DNA binding protein replication protein A (RPA) is also recruited and coats the undamaged strand. XPA recruits a structure-specific endonuclease — the XPF–ERCC1 heterodimer, which is directed to the damaged strand by RPA to create an incision 5′ to the lesion. Once this 'point of no return' is reached, XPG is activated and cuts the damaged strand 3′ to the lesion, which excises the lesion within a 22–30 nucleotide-long strand. The trimeric proliferating cell nuclear antigen (PCNA) ring, which is directly loaded after the 5′ incision by XPF–ERCC1, recruits DNA Pol δ, DNA Pol κ or DNA Pol ε for gap-filling DNA synthesis. Gap filling can begin immediately after the 5′ incision is made. The NER reaction is completed through sealing the final nick by DNA ligase 1 or DNA ligase 3.
Diseases associated
Mutations in GG-NER machinery are responsible for multiple genetic disorders like Xeroderma pigmentosum (XP), which causes severe photosensitivity, high cancer rates in areas of the body exposed to the sun.
Popular Citations
- Petruseva IO, Evdokimov AN, Lavrik OI (2014). “Molecular Mechanism of Global Genome Nucleotide Excision Repair”. Acta Naturae.