Transcription Coupled NER
TC-NER operates when damage to a transcribed DNA strand limits transcription activity. It initiates when RNA polymerase II stalls at a lesion in DNA: the blocked RNA polymerase serves as a damage recognition signal, which replaces the need for the distortion recognition properties of XPC-Rad23B and DDB complexes. CS proteins (CSA and CSB) bind some types of DNA damage instead of XPC-Rad23B.
It was originally dubbed as “preferential repair”, as it was found that UV-induced photoproducts are removed more rapidly from transcribed sequences as compared with nontranscribed DNA, and it appears important to protect cells against UV-light-induced apoptosis.
In case of UV hypersensitvity, in >90% of the UV-induced DNA lesions, the subsequent gap-filling DNA synthesis, measured outside S phase as unscheduled DNA synthesis or UDS, is only marginally affected in TC-NER-deficient cells. Direct monitoring of TC-NER is significantly more laborious; only strand-specific damage removal assays are able to directly measure this NER subpathway.
Damage Recognition
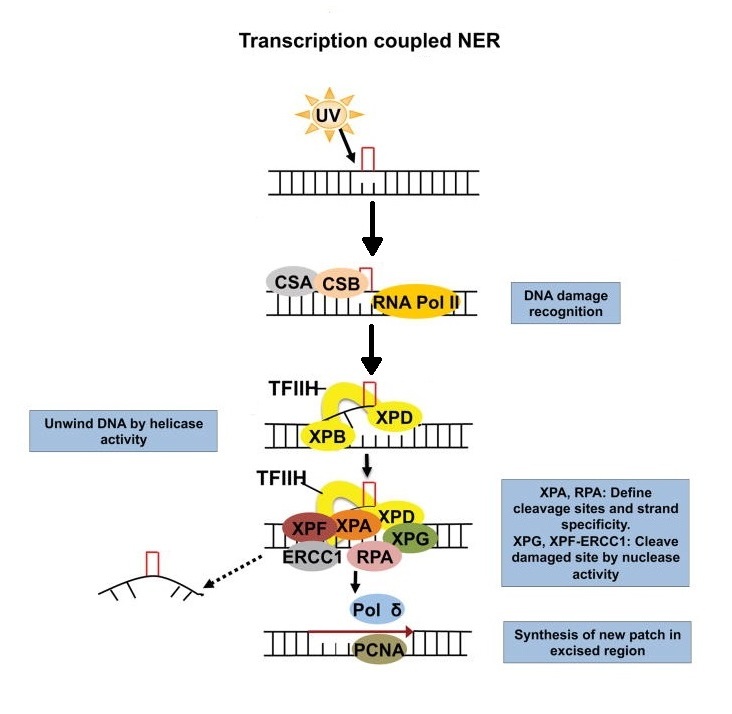
Schematic diagram for mechanisms of transcription coupled nucleotide excision repair (TC-NER) in eukaryotes.
In TC-NER subpathway, damage is indirectly recognized during transcript elongation by the stalling of RNA polymerase II (RNA Pol II) at a lesion. During transcript elongation UV-stimulated scaffold protein A (UVSSA), ubiquitin-specific-processing protease 7 (USP7) and Cockayne syndrome protein CSB transiently interact with RNA Pol II. Upon stalling at a lesion, the affinity of CSB for RNA Pol II increases and the Cockayne syndrome WD repeat protein CSA–CSB complex is formed, which probably results in reverse translocation (backtracking) of RNA Pol II that renders the DNA lesion accessible for repair. After damage recognition, the TFIIH (transcription initiation factor IIH) complex is recruited to the lesion in GG-NER. In NER the XPG structure - specific endonuclease, either associated with TFIIH or separately, binds to the pre-incision NER complex. Upon binding of TFIIH, the CAK (CDK-activating kinase) subcomplex dissociates from the core TFIIH complex. The helicase activity of TFIIH further opens the double helix around the lesion, and 5′–3′ unwinding of the DNA by the TFIIH basal transcription factor complex helicase subunit XPD verifies the existence of lesions with the help of the ATPase activity of the TFIIH XPB subunit and XPA, which binds to single-stranded, chemically altered nucleotides. In this step the single-stranded DNA binding protein replication protein A (RPA) is also recruited and coats the undamaged strand. XPA recruits a structure-specific endonuclease — the XPF–ERCC1 heterodimer, which is directed to the damaged strand by RPA to create an incision 5′ to the lesion. Once this 'point of no return' is reached, XPG is activated and cuts the damaged strand 3′ to the lesion, which excises the lesion within a 22–30 nucleotide-long strand. The trimeric proliferating cell nuclear antigen (PCNA) ring, which is directly loaded after the 5′ incision by XPF–ERCC1, recruits DNA Pol δ, DNA Pol κ or DNA Pol ε for gap-filling DNA synthesis. Gap filling can begin immediately after the 5′ incision is made. The NER reaction is completed through sealing the final nick by DNA ligase 1 or DNA ligase 3.
Diseases associated
TC-NER initiates when RNA polymerase stalls at a lesion in DNA, whereupon protein complexes help move the polymerase backwards. Mutations in TC-NER machinery are responsible for multiple genetic disorders including:
- Cockayne syndrome (CS) causes photosensitivity, mental retardation, progeria-like features, microcephaly in patients.
- Trichothiodystrophy (TTD) causes UV hypersensitvity, ichthyosis, mental/physical retardation in some individuals.
Popular Citations
- Fousteri M, Mullenders LH (2008). "Transcription-coupled nucleotide excision repair in mammalian cells: molecular mechanisms and biological effects". Cell Res. 18.